Abstract
A series of plasmid vectors containing the multiple cloning site (MCS7) of M13mp7 have been constructed. In one of these vectors, a kanamycin resistance marker has been inserted into the centre of the symmetrical MCS7 to produce a restriction site mobilizer (RSM) element. The drug resistance marker can be excised from this vector with any of the restriction enzymes that recognize a site in the RSM flanking sequences to generate an RSM with various sticky ends or blunt ends. These fragments can be used for insertional mutagenesis of any target molecule with compatible restriction sites.
Insertion mutants are selected for their resistance to kanamycin. When the drug resistance marker is removed with PstI, a small in-frame insert can be generated. In addition, two new MCS have been formed that have unique restriction sites by disrupting the symmetrical structure of MCS7. The resulting plasmids pUC8 and pUC9 allow separate double-digested restriction fragments to be cloned in both orientations relative to the lac promoter. The terminal sequences of any DNA cloned into these plasmids can be characterized using the M13 universal primers.
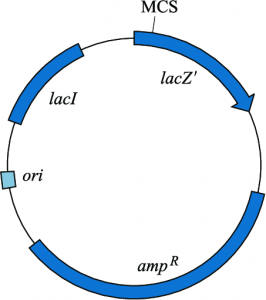
pUC plasmid vectors are used as cloning vectors and belong to the pUC series (named after the place of their initial preparation, ie, the University of California). These plasmids are 2700 bp long and contain
- Ampicillin resistance gene
- Origin of replication derived from pBR322,
- lacZ gene derived from E. coli. Also found within the lac region is a polylinker sequence that has unique restriction sites (identical to those found in phage M13), and
- Multiple Cloning Sites (MCS).
These plasmids, when transformed into an appropriate E.coli strain having lac (eg, JM103, JMI09), and grown in the presence of IPTG (isopropyl thiogalactoside, which behaves like lactose and induces lactose synthesis). of the enzyme galactosidase f3) and X-gal (substrate for the enzyme), will give rise to white or clear colonies.
On the other hand, the pUC that does not have inserts and is transformed into bacteria will have an active lacZ gene and will therefore produce blue colonies, which will make it possible to identify the colonies that have the pUC vector with cloned DNA segments.
As discussed above, in pBR322 and pBR327, the DNA is inserted into a site located in one of the two antibiotic resistance genes, thereby inactivating one of the two resistance genes. The insert-carrying plasmid can be selected for its ability to grow in a medium containing only one of the two antibiotics and for its inability to grow in a medium containing both antibiotics.
On the other hand, plasmids that do not carry an insert grow in media containing one or both antibiotics. Thus, the presence of the lacZ gene in pUC and ampicillin and tetracycline resistance genes in pBR322 and pBR327 allow selection of E. coli colonies transformed with plasmids carrying the desired foreign cloned DNA segment.
pUC19 is a commonly used high copy cloning vector. The vector encodes the N-terminal fragment of β-galactosidase (Lanza), which allows detection of blue/white colonies (i.e. complement a), as well as a pUC origin of replication and an ampicillin resistance gene that allow propagation and selection. in E. coli.
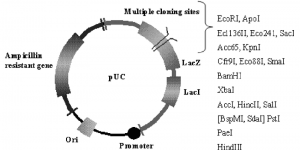
An important feature of pUC plasmids is the blue/white colony screen to detect recombinant plasmids. This selection is based on the inactivation of the Lanza peptide of beta-galactosidase, which is expressed by the vector. Cloning vectors belonging to the pUC family are available in pairs with reverse orders of restriction sites relative to the lacZ promoter. pUC8 and PUC9 form one such pair. Other similar pairs include pUC12 and pUCl3 or pUC18 and pUC19.
Disadvantages of pUC vectors
The basis of genomic sequence analysis is the large-scale cloning and sequencing of shotgun plasmid libraries obtained by assembling sequences from the vast majority of clones.
- Breaches in shotgun libraries and unclonable DNA fragments are quite common. DNA is characterized by high AT content, strong secondary structure, open reading frames, or cis-acting functions (eg, transcriptional promoters or origins of replication).
- In some cases, most notably in AT-rich DNA, the reasons for the difficulty in cloning are not well defined.